Super-resolution analysis of three-dimensional chromosome folding
By combining fluorescent DNA labeling with super-resolution microscopy, a research team has characterized the structure of genomic domains called TADs within which chromatin interactions are enriched. This organization depends on the cohesin complex, which generates 3D interactions, and on the CTCF protein, which prevents contacts between neighboring TADs. TADs are further subdivided into "nanodomains" which depend on the chromatin epigenetic profile. These results are published in the journal Nature Genetics.
The study of the three-dimensional organization of the genome reveals functional relationships between chromosome structure and gene regulation. Within the nucleus of eukaryotic cells, the genome is folded hierarchically. At the smallest scale, the DNA molecule wraps around histone proteins, forming nucleosomes and the primary structure of the chromatin fiber. On the highest scale, each chromosome occupies its own territory within the nucleus. Between these two extremes, the physical organization of chromosomes remains poorly characterized, while it is at this scale that most functional processes, such as DNA replication and RNA transcription occur. During the last decade, the use of a sophisticated molecular biology method called Hi-C, which allows the mapping of all chromosomal contacts, has revealed the presence of genomic domains of the order of Megabase (one million of base pairs) enriched in interactions: the so-called “Topologically Associating Domains” or TADs. Different studies have shed light on the functional role of TADs in gene regulation, in particular through their ability to favor long-distance contacts between gene promoters (transcription regulation sites) and their regulatory sequences such as “enhancers” when enhancers are located in the same TAD. The disruption of TADs is associated with developmental diseases or certain cancers. However, the Hi-C method generating chromosomal interaction maps is obtained from large populations of cells. The direct characterization of the structure of these domains in individual cells is essential in order to understand how the genome is physically folded into TADs.
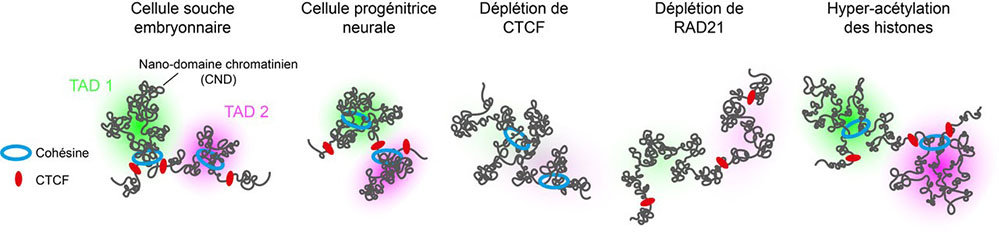
In the present study, a DNA-specific labeling technique using fluorescent oligonucleotides called "Oligopaint Fluorescent in situ hybridization" was combined with super-resolution microscopy in order to study the structure of TADs with a spatial resolution in the order of a hundred nanometers (one nanometer is one billionths of a meter). The analysis of many chromosomal regions in mouse embryonic stem cells has revealed a great structural heterogeneity of TADs. Despite this variability, the majority of cells show enriched contacts within a TAD, compared to neighboring TADs. These results are therefore consistent with the proposed role of TADs in the physical definition of functional genomic domains. Moreover, their spatial segregation is exacerbated in neural progenitor cells compared to stem cells, suggesting that the folding of chromosomes into TADs is intensified during cell differentiation.
The researchers also studied the effects of depleting the CTCF protein, which is normally located at the borders between TADs, as well as of the RAD21 subunit of the cohesin complex. In the absence of either of these proteins, preferential chromatin contacts within TADs are abolished, confirming previous observations using the Hi-C assay. However, the disappearance of TADs occurs through two distinct mechanisms. The depletion of CTCF leads to increased interactions between neighboring TADs, highlighting its role as a barrier protein. The depletion of RAD21, on the other hand, leads to a loss of intra-TAD contacts, revealing its general role in the generation of chromatin interactions. These results, consistent with a model of TAD formation by loop extrusion, indicate that the cohesin complex is responsible for the enrichment of chromatin contacts, while CTCF, by defining the boundaries, constrains the action of cohesin within TADs.
Finally, the use of super-resolution microscopy revealed the presence of substructures within TADs, that were defined as "chromatin nano-domains" or CNDs. Their organization depends on the chromatin epigenetic state. Indeed, the induction of histone hyper-acetylation disrupts the formation of CNDs, presumably by antagonizing interactions between nucleosomes. These data therefore suggest that CNDs are formed by aggregation of nucleosomes.
This study thus characterizes the physical organization of TADs in single cells, clarifies the roles of CTCF and cohesin in their formation, and reveals the existence of CNDs, which represent a new level of structural organization at an intermediate between nucleosomes and TADs. Future studies should provide a better understanding of the molecular mechanisms involved in CND formation as well as their functional roles in genome regulation.
Figure
Representation of chromosome within TADs. TAD boundaries are occupied by the CTCF protein, while the cohesin complex generates chromatin interactions within TADs. TADs are subdivided into chromatin nanomains (CNDs), which are formed by nucleosome aggregation. The spatial segregation of TADs is enhanced during embryonic stem cell differentiation into neural progenitor cells. Upon depletion of CTCF, the cohesin complex can continue to act across the boundaries between TADs, thus inducing ectopic contacts between neighboring TADs. Upon RAD21 depletion, chromatin interactions generated within TADs are abolished. Upon induction of histone the hyperacetylation, TADs remain spatially separated but their internal organization into CNDs is perturbed.
Learn more
Regulation of single-cell genome organization into TADs and chromatin nanodomains.
Szabo Q, Donjon A, Jerković I, Papadopoulos GL, Cheutin T, Bonev B, Nora EP, Bruneau BG, Bantignies F, Cavalli G.
Nat Genet. 2020 Oct 19. doi: 10.1038/s41588-020-00716-8.
Contact
Laboratory
Institut de Génétique Humaine (IGH) - (CNRS/Université de Montpellier)
141, rue de la Cardonille
34396 Montpellier - Cedex 5, France